library(rfintext)
# devtools::install_github("StranMax/rfinstats")
library(rfinstats)
library(tidymodels)
library(topicmodels)
library(cowplot)
library(dplyr)
library(tidytext)
library(ggplot2)
library(quanteda)
library(forcats)
library(purrr)
library(future) # Parallel processing back-end
library(furrr) # Parallel processing front-end with future_ functions
plan(multisession, workers = availableCores(logical = FALSE) - 1)
dtm <- aspol |>
preprocess_corpus(kunta) |>
corpus_to_dtm(kunta, LEMMA)
y <- rfinstats::taantuvat |>
filter(kunta %in% unique(aspol$kunta)) |>
mutate(luokka = if_else(suht_muutos_2010_2022 > 0, "kasvava", "taantuva"))
y
#> # A tibble: 66 × 5
#> kunta vaesto kokmuutos_2010_2022 suht_muutos_2010_2022 luokka
#> <chr> <int> <int> <dbl> <chr>
#> 1 Enontekiö 1876 -71 -3.78 taantuva
#> 2 Espoo 247970 60944 24.6 kasvava
#> 3 Eura 12507 -1278 -10.2 taantuva
#> 4 Hartola 3355 -814 -24.3 taantuva
#> 5 Hattula 9657 -266 -2.75 taantuva
#> 6 Helsinki 588549 80678 13.7 kasvava
#> 7 Huittinen 10663 -955 -8.96 taantuva
#> 8 Hyvinkää 45489 1527 3.36 kasvava
#> 9 Hämeenlinna 66829 1588 2.38 kasvava
#> 10 Iitti 7005 -557 -7.95 taantuva
#> # ℹ 56 more rows
optimal_params <- lda_models |>
filter(K == 18) |>
slice_max(mean_coherence, n = 1)
optimal_params
#> # A tibble: 1 × 3
#> K S mean_coherence
#> <int> <int> <dbl>
#> 1 18 5883911 -9.39
# optimal_k <- c(5, 15, 18, 21)
# optimal_k <- c(5, 8, 11, 14, 19)
# optimal_k <- c(5, 7, 10, 15, 21) # Visually selected based on mean topic coherence (see article 3)
ptm <- proc.time()
lda_models <- optimal_params |>
# slice_max(mean_coherence, n = 1) |>
mutate(
# LDA model
topic_model = future_map2(
K, S, \(k, s) {
LDA(dtm, k = k, control = list(seed = S))
}, .options = furrr_options(seed = NULL)
),
# Beta matrix
beta = map(
topic_model, \(x) tidy(x, matrix = "beta")
),
# Theta matrix (gamma)
theta = map(
topic_model, \(x) {
tidy(x, matrix = "gamma") |>
filter(document %in% y$kunta) |>
rename(kunta = document, theta = gamma)
}
)
)
proc.time() - ptm
#> user system elapsed
#> 1.166 0.047 45.084
lda_models
#> # A tibble: 1 × 6
#> K S mean_coherence topic_model beta theta
#> <int> <int> <dbl> <list> <list> <list>
#> 1 18 5883911 -9.39 <LDA_VEM> <tibble [54,684 × 3]> <tibble>
lda_models <- lda_models |>
mutate(theta = map(theta, \(df) {
df |>
left_join(y) |>
select(aihe = topic, theta, luokka) |>
mutate(luokka = factor(luokka))
}))
#> Joining with `by = join_by(kunta)`
lda_models
#> # A tibble: 1 × 6
#> K S mean_coherence topic_model beta theta
#> <int> <int> <dbl> <list> <list> <list>
#> 1 18 5883911 -9.39 <LDA_VEM> <tibble [54,684 × 3]> <tibble>We calculate null distribution of t-values and observed t-value:
ptm <- proc.time()
t_distributions <- lda_models |>
select(K, theta) |>
unnest(theta) |>
nest(data = c(theta, luokka)) |>
mutate(
null_dist = future_map(data, \(df) {
# t-distribution under null hypothesis
df |>
specify(theta ~ luokka) |>
hypothesize(null = "independence") |>
generate(reps = 100000, type = "permute") |>
calculate("diff in means", order = c("kasvava", "taantuva"))
}, .options = furrr_options(seed = TRUE)),
# t-value for estimation
t_val = map_dbl(data, \(df) {
df |>
specify(theta ~ luokka) |>
calculate("diff in means", order = c("kasvava", "taantuva")) |>
pull(stat)
})
)
proc.time() - ptm
#> user system elapsed
#> 9.044 0.218 164.645
t_distributions
#> # A tibble: 18 × 5
#> K aihe data null_dist t_val
#> <int> <int> <list> <list> <dbl>
#> 1 18 1 <tibble [66 × 2]> <infer [100,000 × 2]> 0.0443
#> 2 18 2 <tibble [66 × 2]> <infer [100,000 × 2]> -0.107
#> 3 18 3 <tibble [66 × 2]> <infer [100,000 × 2]> -0.0524
#> 4 18 4 <tibble [66 × 2]> <infer [100,000 × 2]> -0.0456
#> 5 18 5 <tibble [66 × 2]> <infer [100,000 × 2]> -0.285
#> 6 18 6 <tibble [66 × 2]> <infer [100,000 × 2]> 0.00608
#> 7 18 7 <tibble [66 × 2]> <infer [100,000 × 2]> 0.0424
#> 8 18 8 <tibble [66 × 2]> <infer [100,000 × 2]> -0.00678
#> 9 18 9 <tibble [66 × 2]> <infer [100,000 × 2]> 0.135
#> 10 18 10 <tibble [66 × 2]> <infer [100,000 × 2]> 0.0232
#> 11 18 11 <tibble [66 × 2]> <infer [100,000 × 2]> 0.0585
#> 12 18 12 <tibble [66 × 2]> <infer [100,000 × 2]> -0.0400
#> 13 18 13 <tibble [66 × 2]> <infer [100,000 × 2]> 0.0117
#> 14 18 14 <tibble [66 × 2]> <infer [100,000 × 2]> 0.130
#> 15 18 15 <tibble [66 × 2]> <infer [100,000 × 2]> 0.0173
#> 16 18 16 <tibble [66 × 2]> <infer [100,000 × 2]> -0.0809
#> 17 18 17 <tibble [66 × 2]> <infer [100,000 × 2]> 0.0663
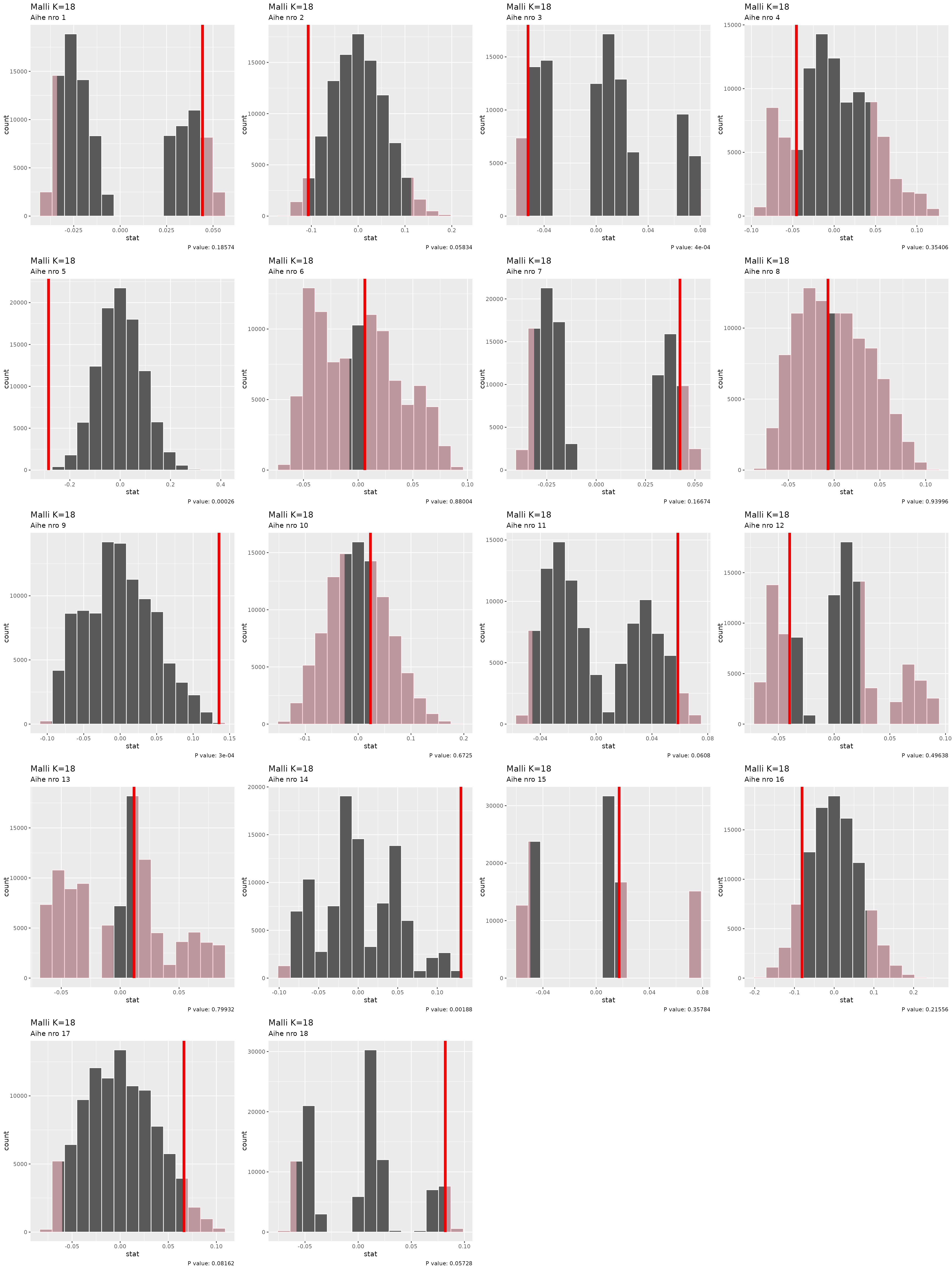
#> 18 18 18 <tibble [66 × 2]> <infer [100,000 × 2]> 0.0819Next we calculate p-values:
t_distributions <- t_distributions |>
mutate(
p_value = pmap_dbl(select(t_distributions, null_dist, t_val), \(null_dist, t_val) {
null_dist |>
get_p_value(obs_stat = t_val, direction = "two-sided") |>
pull(p_value)
})
)
t_distributions
#> # A tibble: 18 × 6
#> K aihe data null_dist t_val p_value
#> <int> <int> <list> <list> <dbl> <dbl>
#> 1 18 1 <tibble [66 × 2]> <infer [100,000 × 2]> 0.0443 0.186
#> 2 18 2 <tibble [66 × 2]> <infer [100,000 × 2]> -0.107 0.0583
#> 3 18 3 <tibble [66 × 2]> <infer [100,000 × 2]> -0.0524 0.0004
#> 4 18 4 <tibble [66 × 2]> <infer [100,000 × 2]> -0.0456 0.354
#> 5 18 5 <tibble [66 × 2]> <infer [100,000 × 2]> -0.285 0.00026
#> 6 18 6 <tibble [66 × 2]> <infer [100,000 × 2]> 0.00608 0.880
#> 7 18 7 <tibble [66 × 2]> <infer [100,000 × 2]> 0.0424 0.167
#> 8 18 8 <tibble [66 × 2]> <infer [100,000 × 2]> -0.00678 0.940
#> 9 18 9 <tibble [66 × 2]> <infer [100,000 × 2]> 0.135 0.0003
#> 10 18 10 <tibble [66 × 2]> <infer [100,000 × 2]> 0.0232 0.672
#> 11 18 11 <tibble [66 × 2]> <infer [100,000 × 2]> 0.0585 0.0608
#> 12 18 12 <tibble [66 × 2]> <infer [100,000 × 2]> -0.0400 0.496
#> 13 18 13 <tibble [66 × 2]> <infer [100,000 × 2]> 0.0117 0.799
#> 14 18 14 <tibble [66 × 2]> <infer [100,000 × 2]> 0.130 0.00188
#> 15 18 15 <tibble [66 × 2]> <infer [100,000 × 2]> 0.0173 0.358
#> 16 18 16 <tibble [66 × 2]> <infer [100,000 × 2]> -0.0809 0.216
#> 17 18 17 <tibble [66 × 2]> <infer [100,000 × 2]> 0.0663 0.0816
#> 18 18 18 <tibble [66 × 2]> <infer [100,000 × 2]> 0.0819 0.0573
plot_grid(plotlist = pmap(select(t_distributions, -data), \(K, aihe, null_dist, t_val, p_value) {
visualise(null_dist) +
shade_p_value(obs_stat = t_val, direction = "two_sided") +
labs(subtitle = paste0("Aihe nro ", aihe),
title = paste0("Malli K=", K),
caption = paste0("P value: ", p_value))
}),
ncol = 4
)
# for (i in unique(theta_matrix$model)) {
# print(
# theta_matrix |>
# filter(model == i) |>
# slice_max(gamma, by = document) |>
# arrange(topic) |>
# left_join(taantuvat, by = join_by("document" == "kunta")) |>
# mutate(topic = factor(topic)) |>
# ggplot(aes(x = topic, y = suht_muutos_2010_2022, colour = gamma)) +
# geom_point() +
# scale_colour_viridis_c(option = "A") +
# labs(title = "Most likely topic per document", subtitle = paste0("Model_", i))
# )
# }
# for (i in unique(theta_matrix$model)) {
# print(
# theta_matrix |>
# filter(model == i) |>
# left_join(taantuvat, by = join_by("document" == "kunta")) |>
# summarise(mean_gamma = mean(gamma), gamma_median = median(gamma), .by = c(luokka, topic)) |>
# mutate(topic = factor(topic)) |>
# ggplot() +
# geom_point(aes(x = topic, y = mean_gamma, colour = luokka), shape = 16) +
# geom_point(aes(x = topic, y = gamma_median, colour = luokka), shape = 17) +
# scale_colour_viridis_d(option = "A") +
# labs(title = "Most likely topic per class", subtitle = paste0("Model_", i))
# )
# }
# for (i in unique(theta_matrix$model)) {
# print(
# theta_matrix |>
# filter(model == i) |>
# left_join(taantuvat, by = join_by("document" == "kunta")) |>
# mutate(luokka = if_else(suht_muutos_2010_2022 > 0, "Kasvava", "Taantuva"),
# topic = factor(as.integer(topic))) |>
# summarise(gamma_sum = sum(gamma),
# gamma_mean = mean(gamma),
# gamma_median = median(gamma),
# .by = c(luokka, topic)) |>
# ggplot() +
# geom_tile(aes(x = topic, y = luokka, fill = gamma_mean)) +
# scale_fill_viridis_c(option = "A") +
# labs(title = "Common topics per class", subtitle = paste0("Model_", i))
# )
# }
# for (i in unique(theta_matrix$model)) {
# print(
# theta_matrix |>
# filter(model == i) |>
# left_join(taantuvat, by = join_by("document" == "kunta")) |>
# mutate(luokka = if_else(suht_muutos_2010_2022 > 0, "Kasvava", "Taantuva"),
# topic = factor(as.integer(topic))) |>
# summarise(gamma_sum = sum(gamma),
# gamma_mean = mean(gamma),
# gamma_median = median(gamma),
# .by = c(luokka, topic)) |>
# ggplot() +
# geom_tile(aes(x = topic, y = luokka, fill = gamma_median)) +
# scale_fill_viridis_c(option = "A") +
# labs(title = "Common topics per class", subtitle = paste0("Model_", i))
# )
# }