library(rfintext)
# devtools::install_github("StranMax/rfinstats")
library(rfinstats)
library(dplyr)
library(tidyr)
library(tidytext)
library(xgboost)
library(caret)
library(topicmodels)
library(quanteda)
library(forcats)
library(purrr)
library(future)
library(furrr)
plan(multisession, workers = availableCores(logical = FALSE) - 1)
y <- rfinstats::taantuvat |>
filter(kunta %in% unique(aspol$kunta)) |>
mutate(luokka = factor(if_else(suht_muutos_2010_2022 > 0, "kasvava", "taantuva")))
y
#> # A tibble: 66 × 5
#> kunta vaesto kokmuutos_2010_2022 suht_muutos_2010_2022 luokka
#> <chr> <int> <int> <dbl> <fct>
#> 1 Enontekiö 1876 -71 -3.78 taantuva
#> 2 Espoo 247970 60944 24.6 kasvava
#> 3 Eura 12507 -1278 -10.2 taantuva
#> 4 Hartola 3355 -814 -24.3 taantuva
#> 5 Hattula 9657 -266 -2.75 taantuva
#> 6 Helsinki 588549 80678 13.7 kasvava
#> 7 Huittinen 10663 -955 -8.96 taantuva
#> 8 Hyvinkää 45489 1527 3.36 kasvava
#> 9 Hämeenlinna 66829 1588 2.38 kasvava
#> 10 Iitti 7005 -557 -7.95 taantuva
#> 11 Imatra 28540 -3468 -12.2 taantuva
#> 12 Inkoo 5546 -225 -4.06 taantuva
#> 13 Joensuu 73305 4809 6.56 kasvava
#> 14 Juva 6962 -1295 -18.6 taantuva
#> 15 Järvenpää 38680 6922 17.9 kasvava
#> 16 Kaarina 30911 5088 16.5 kasvava
#> 17 Kalajoki 12562 -205 -1.63 taantuva
#> 18 Kauniainen 8689 1667 19.2 kasvava
#> 19 Kemiönsaari 7191 -749 -10.4 taantuva
#> 20 Kerava 34282 3843 11.2 kasvava
#> # ℹ 46 more rows
dtm <- aspol |>
preprocess_corpus(kunta) |>
corpus_to_dtm(kunta, LEMMA)Use 13 topics, and a few random seeds.
optimal_params <- lda_models |>
filter(K == 18) |>
slice_max(mean_coherence, n = 1)
optimal_params
#> # A tibble: 1 × 3
#> K S mean_coherence
#> <int> <int> <dbl>
#> 1 18 5883911 -9.39
# optimal_k <- c(5, 7, 10, 15, 21) # Visually selected based on mean topic coherence (see article 3)
ptm <- proc.time()
lda_models <- optimal_params |>
mutate(
# LDA models
topic_model = future_map2(
K, S, \(k, s) {
LDA(dtm, k = k, control = list(seed = s))
}, .options = furrr_options(seed = NULL) # use with furrr::future_map2()
),
# Theta matrix (gamma)
theta = map(
topic_model, \(x) {
tidy(x, matrix = "gamma") |>
rename(theta = gamma) |>
filter(document %in% y$kunta) |>
cast_dfm(document = document, term = topic, value = theta)
}
)
)
proc.time() - ptm
#> user system elapsed
#> 1.259 0.046 45.006
lda_models
#> # A tibble: 1 × 5
#> K S mean_coherence topic_model theta
#> <int> <int> <dbl> <list> <list>
#> 1 18 5883911 -9.39 <LDA_VEM> <dfm[,18]>Divide data set to training and test parts:
trainidx <- createDataPartition(y$suht_muutos_2010_2022, p = .7,
list = FALSE,
times = 1)
lda_models <- lda_models |>
mutate(
train = map(theta, \(x) {
dfm_subset(x, docname_ %in% y$kunta[trainidx])
}),
test = map(theta, \(x) {
dfm_subset(x, docname_ %in% y$kunta[-trainidx])
})
)
lda_models
#> # A tibble: 1 × 7
#> K S mean_coherence topic_model theta train test
#> <int> <int> <dbl> <list> <list> <list> <list>
#> 1 18 5883911 -9.39 <LDA_VEM> <dfm[,18]> <dfm[,18]> <dfm[,18]>
lda_models <- lda_models |>
mutate(
xgb_train = map(train, \(train) {
xgb.DMatrix(data = train, label = as.integer(y$luokka[trainidx]) - 1) # Class levels to integers (starting from 0)
}),
xgb_test = map(test, \(test) {
xgb.DMatrix(data = test, label = as.integer(y$luokka[-trainidx]) - 1) # Class levels to integers (starting from 0)
})
)
lda_models
#> # A tibble: 1 × 9
#> K S mean_coherence topic_model theta train test
#> <int> <int> <dbl> <list> <list> <list> <list>
#> 1 18 5883911 -9.39 <LDA_VEM> <dfm[,18]> <dfm[,18]> <dfm[,18]>
#> # ℹ 2 more variables: xgb_train <list>, xgb_test <list>Define set of parameters for hyper parameter tuning: Only small subset of possible values defined here to cut down processing time for package website.
gs <- tidyr::expand_grid(
booster = "gbtree",
eta = seq(0.01, 0.1, by = 0.3),
max_depth = seq(2, 6, by = 1),
gamma = seq(0, 4, by = 2),
subsample = seq(0.5, 1, by = 0.25),
colsample_bylevel = seq(0.5, 1, by = 0.25),
nrounds = seq(5, 55, by = 25),
# objective = "reg:squarederror",
objective = "binary:logistic",
num_parallel_tree = 2,
)
gs
#> # A tibble: 405 × 9
#> booster eta max_depth gamma subsample colsample_bylevel nrounds objective num_parallel_tree
#> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <chr> <dbl>
#> 1 gbtree 0.01 2 0 0.5 0.5 5 binary:logistic 2
#> 2 gbtree 0.01 2 0 0.5 0.5 30 binary:logistic 2
#> 3 gbtree 0.01 2 0 0.5 0.5 55 binary:logistic 2
#> 4 gbtree 0.01 2 0 0.5 0.75 5 binary:logistic 2
#> 5 gbtree 0.01 2 0 0.5 0.75 30 binary:logistic 2
#> 6 gbtree 0.01 2 0 0.5 0.75 55 binary:logistic 2
#> 7 gbtree 0.01 2 0 0.5 1 5 binary:logistic 2
#> 8 gbtree 0.01 2 0 0.5 1 30 binary:logistic 2
#> 9 gbtree 0.01 2 0 0.5 1 55 binary:logistic 2
#> 10 gbtree 0.01 2 0 0.75 0.5 5 binary:logistic 2
#> 11 gbtree 0.01 2 0 0.75 0.5 30 binary:logistic 2
#> 12 gbtree 0.01 2 0 0.75 0.5 55 binary:logistic 2
#> 13 gbtree 0.01 2 0 0.75 0.75 5 binary:logistic 2
#> 14 gbtree 0.01 2 0 0.75 0.75 30 binary:logistic 2
#> 15 gbtree 0.01 2 0 0.75 0.75 55 binary:logistic 2
#> 16 gbtree 0.01 2 0 0.75 1 5 binary:logistic 2
#> 17 gbtree 0.01 2 0 0.75 1 30 binary:logistic 2
#> 18 gbtree 0.01 2 0 0.75 1 55 binary:logistic 2
#> 19 gbtree 0.01 2 0 1 0.5 5 binary:logistic 2
#> 20 gbtree 0.01 2 0 1 0.5 30 binary:logistic 2
#> # ℹ 385 more rowsAdd combination of lda-models to hyper parameter set:
xgb_models <- expand_grid(select(lda_models, K), gs) |>
left_join(select(lda_models, K, data = xgb_train, test_data = xgb_test))
#> Joining with `by = join_by(K)`
xgb_models
#> # A tibble: 405 × 12
#> K booster eta max_depth gamma subsample colsample_bylevel nrounds
#> <int> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 18 gbtree 0.01 2 0 0.5 0.5 5
#> 2 18 gbtree 0.01 2 0 0.5 0.5 30
#> 3 18 gbtree 0.01 2 0 0.5 0.5 55
#> 4 18 gbtree 0.01 2 0 0.5 0.75 5
#> 5 18 gbtree 0.01 2 0 0.5 0.75 30
#> 6 18 gbtree 0.01 2 0 0.5 0.75 55
#> 7 18 gbtree 0.01 2 0 0.5 1 5
#> 8 18 gbtree 0.01 2 0 0.5 1 30
#> 9 18 gbtree 0.01 2 0 0.5 1 55
#> 10 18 gbtree 0.01 2 0 0.75 0.5 5
#> # ℹ 395 more rows
#> # ℹ 4 more variables: objective <chr>, num_parallel_tree <dbl>, data <list>,
#> # test_data <list>Now we are ready to train models:
ptm <- proc.time()
xgb_models <- xgb_models |>
mutate(model = pmap(select(xgb_models, -K, -test_data), xgb.train)) # future_pmap does not work here!
proc.time() - ptm
#> user system elapsed
#> 12.847 0.831 4.102
xgb_models
#> # A tibble: 405 × 13
#> K booster eta max_depth gamma subsample colsample_bylevel nrounds
#> <int> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 18 gbtree 0.01 2 0 0.5 0.5 5
#> 2 18 gbtree 0.01 2 0 0.5 0.5 30
#> 3 18 gbtree 0.01 2 0 0.5 0.5 55
#> 4 18 gbtree 0.01 2 0 0.5 0.75 5
#> 5 18 gbtree 0.01 2 0 0.5 0.75 30
#> 6 18 gbtree 0.01 2 0 0.5 0.75 55
#> 7 18 gbtree 0.01 2 0 0.5 1 5
#> 8 18 gbtree 0.01 2 0 0.5 1 30
#> 9 18 gbtree 0.01 2 0 0.5 1 55
#> 10 18 gbtree 0.01 2 0 0.75 0.5 5
#> # ℹ 395 more rows
#> # ℹ 5 more variables: objective <chr>, num_parallel_tree <dbl>, data <list>,
#> # test_data <list>, model <list>Calculate errors for all models:
xgb_models <- xgb_models |>
mutate(
error = map2_dbl(model, test_data, \(model, test_data) {
label = xgboost::getinfo(test_data, "label")
pred <- stats::predict(model, test_data)
err <- as.numeric(sum(as.integer(pred > 0.5) != label))/length(label)
err
})
)
xgb_models
#> # A tibble: 405 × 14
#> K booster eta max_depth gamma subsample colsample_bylevel nrounds
#> <int> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 18 gbtree 0.01 2 0 0.5 0.5 5
#> 2 18 gbtree 0.01 2 0 0.5 0.5 30
#> 3 18 gbtree 0.01 2 0 0.5 0.5 55
#> 4 18 gbtree 0.01 2 0 0.5 0.75 5
#> 5 18 gbtree 0.01 2 0 0.5 0.75 30
#> 6 18 gbtree 0.01 2 0 0.5 0.75 55
#> 7 18 gbtree 0.01 2 0 0.5 1 5
#> 8 18 gbtree 0.01 2 0 0.5 1 30
#> 9 18 gbtree 0.01 2 0 0.5 1 55
#> 10 18 gbtree 0.01 2 0 0.75 0.5 5
#> # ℹ 395 more rows
#> # ℹ 6 more variables: objective <chr>, num_parallel_tree <dbl>, data <list>,
#> # test_data <list>, model <list>, error <dbl>Smallest errors appear to be 0.1111111
xgb_models |> arrange(error)
#> # A tibble: 405 × 14
#> K booster eta max_depth gamma subsample colsample_bylevel nrounds
#> <int> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 18 gbtree 0.01 5 4 0.5 0.75 5
#> 2 18 gbtree 0.01 4 0 0.75 1 30
#> 3 18 gbtree 0.01 4 2 0.5 0.75 5
#> 4 18 gbtree 0.01 4 2 0.75 0.75 5
#> 5 18 gbtree 0.01 6 0 0.75 0.5 5
#> 6 18 gbtree 0.01 2 2 1 0.5 30
#> 7 18 gbtree 0.01 2 4 0.75 0.5 5
#> 8 18 gbtree 0.01 3 0 0.75 0.5 30
#> 9 18 gbtree 0.01 3 0 0.75 0.75 55
#> 10 18 gbtree 0.01 3 0 1 0.5 55
#> # ℹ 395 more rows
#> # ℹ 6 more variables: objective <chr>, num_parallel_tree <dbl>, data <list>,
#> # test_data <list>, model <list>, error <dbl>
xgb_models |>
ggplot(aes(x = error)) +
geom_boxplot() +
coord_flip() +
facet_wrap(~K, ncol = 5)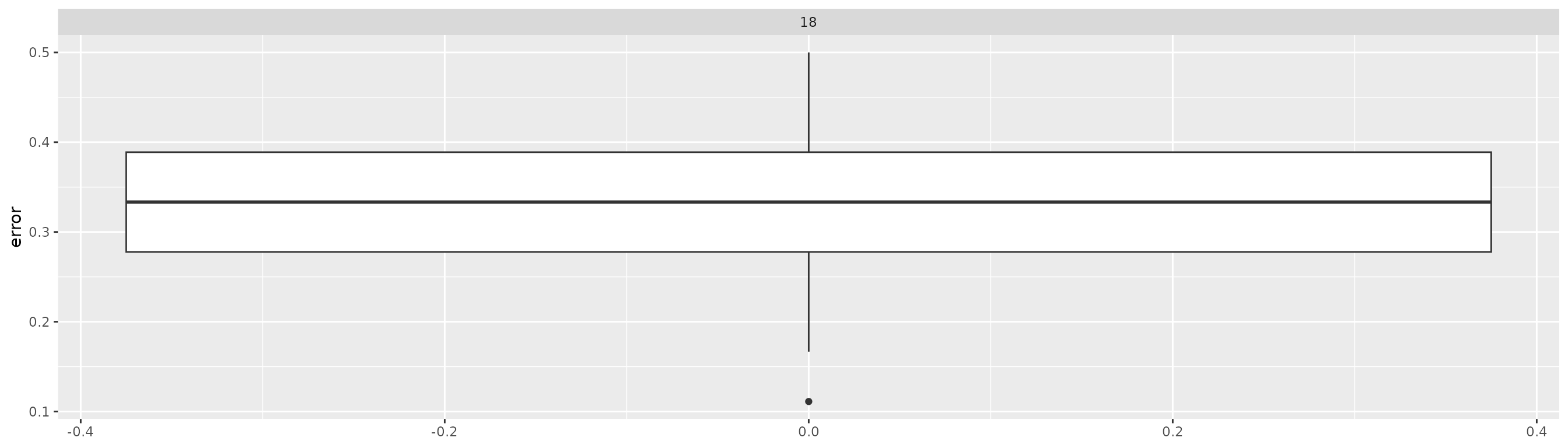
xgb_models |>
ggplot(aes(x = error)) +
geom_boxplot() +
coord_flip() +
facet_wrap(~max_depth, ncol = 6)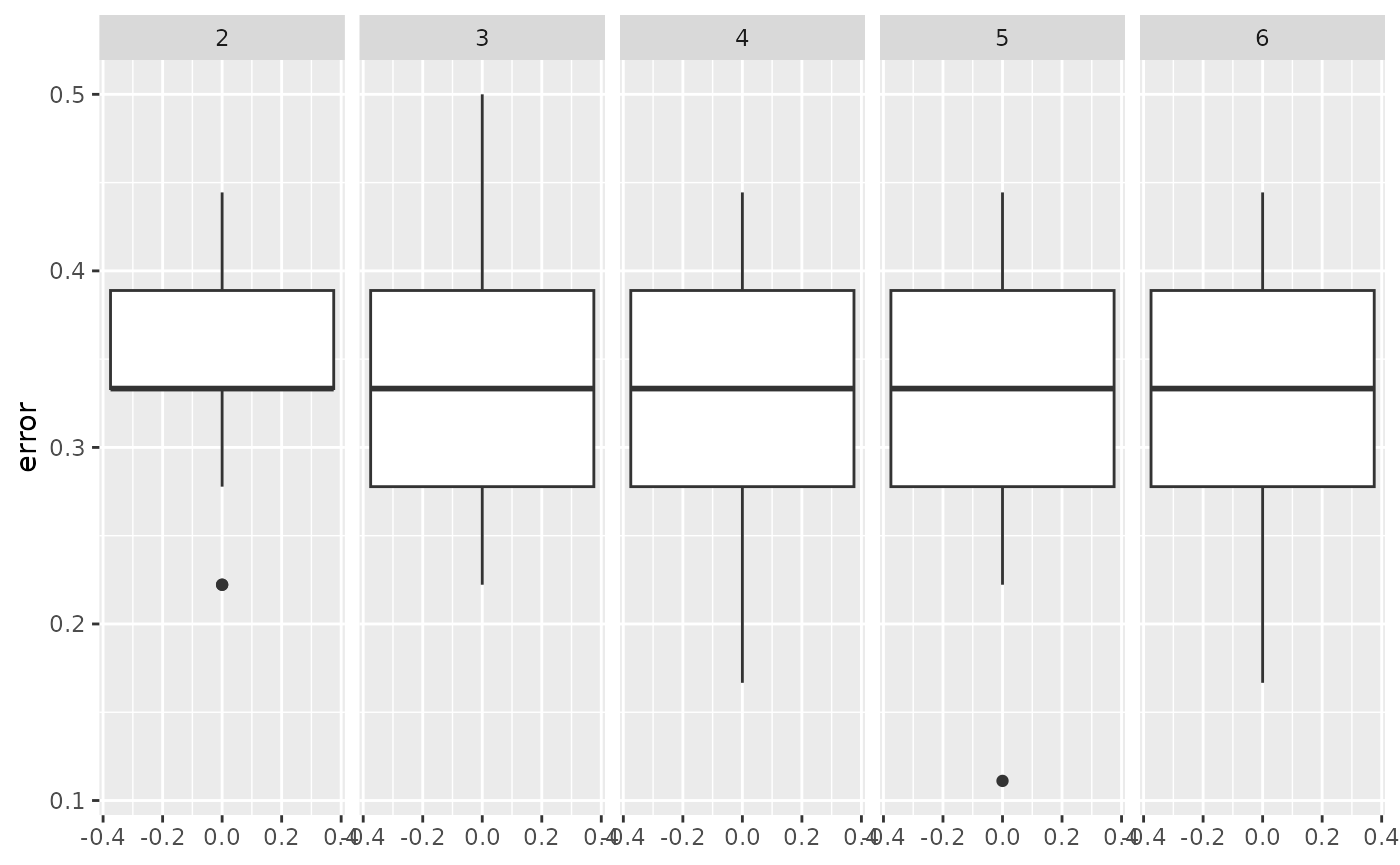
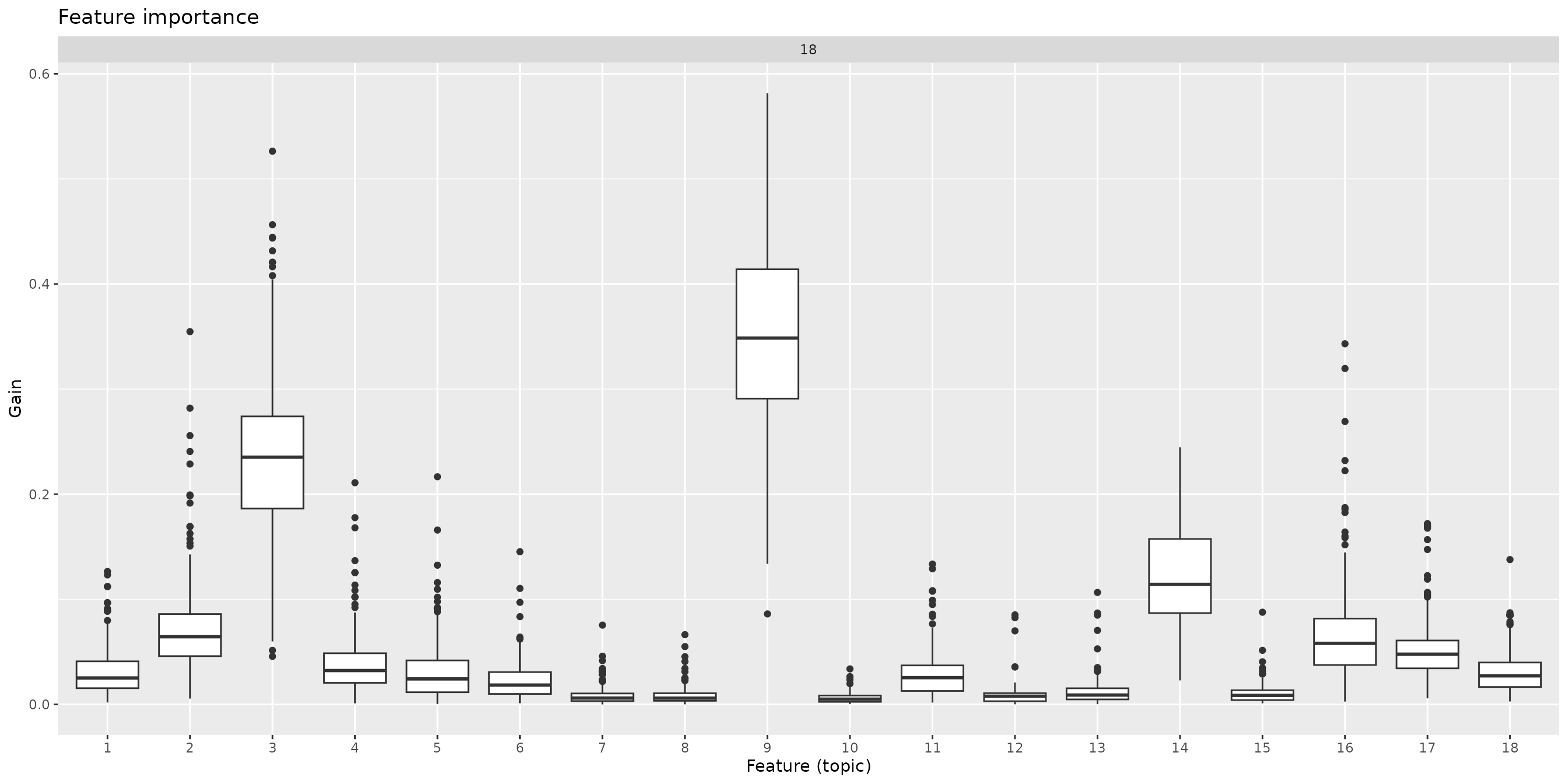
Feature importance:
xgb_models <- xgb_models |>
mutate(
feat_importance = map2(data, model, \(data, model) {
xgb.importance(feature_names = colnames(data),
model = model)
})
)
xgb_models
#> # A tibble: 405 × 15
#> K booster eta max_depth gamma subsample colsample_bylevel nrounds
#> <int> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 18 gbtree 0.01 2 0 0.5 0.5 5
#> 2 18 gbtree 0.01 2 0 0.5 0.5 30
#> 3 18 gbtree 0.01 2 0 0.5 0.5 55
#> 4 18 gbtree 0.01 2 0 0.5 0.75 5
#> 5 18 gbtree 0.01 2 0 0.5 0.75 30
#> 6 18 gbtree 0.01 2 0 0.5 0.75 55
#> 7 18 gbtree 0.01 2 0 0.5 1 5
#> 8 18 gbtree 0.01 2 0 0.5 1 30
#> 9 18 gbtree 0.01 2 0 0.5 1 55
#> 10 18 gbtree 0.01 2 0 0.75 0.5 5
#> # ℹ 395 more rows
#> # ℹ 7 more variables: objective <chr>, num_parallel_tree <dbl>, data <list>,
#> # test_data <list>, model <list>, error <dbl>, feat_importance <list>
xgb_models |>
select(K, feat_importance) |>
unnest(feat_importance) |>
mutate(Feature = factor(as.integer(Feature))) |>
ggplot(aes(x = Feature, y = Gain)) +
geom_boxplot() +
labs(title = "Feature importance", x = "Feature (topic)") +
facet_wrap(~K, scales = "free_x")